We specialize in developing technologies in the space of genome editing and bifunctional molecules, and applying those technologies to beta-cell biology.
CRISPR-associated nucleases (e.g., Cas9) are programmable RNA-guided nuclease that continue to furnish disruptive technologies, including those for genome-, epigenome- and transcriptome-editing, and transcription activation/repression. Precision control lies at the heart of powerful technologies and CRISPR-based technologies are no exceptions—they require multidimensional control over half-life, dosage, and spatiotemporal (See review and TEDMED talk here). We develop CRISPR controllers (activators, inhibitors, and degraders) to elevate these technologies to their fullest potential, pivoting on specific applications in beta-cell engineering (see below).
Activators
With the innumerable negative consequences of elevated and prolonged activity of Cas9 in a cell (e.g., off-target editing, genotoxicity), many applications would benefit from a default switched-off state for Cas9-based technologies can be activated at precise times and with dose control. Our lab has developed chemical and photo-chemical activators to control Cas9-based technologies in cells and organisms (e.g., gene drives).
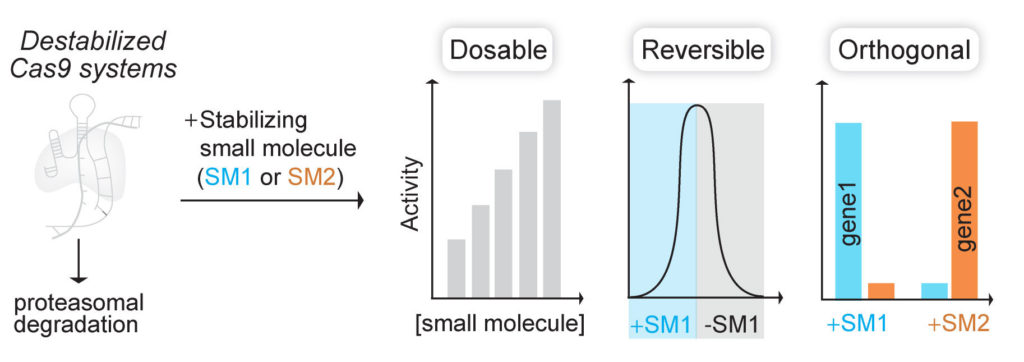
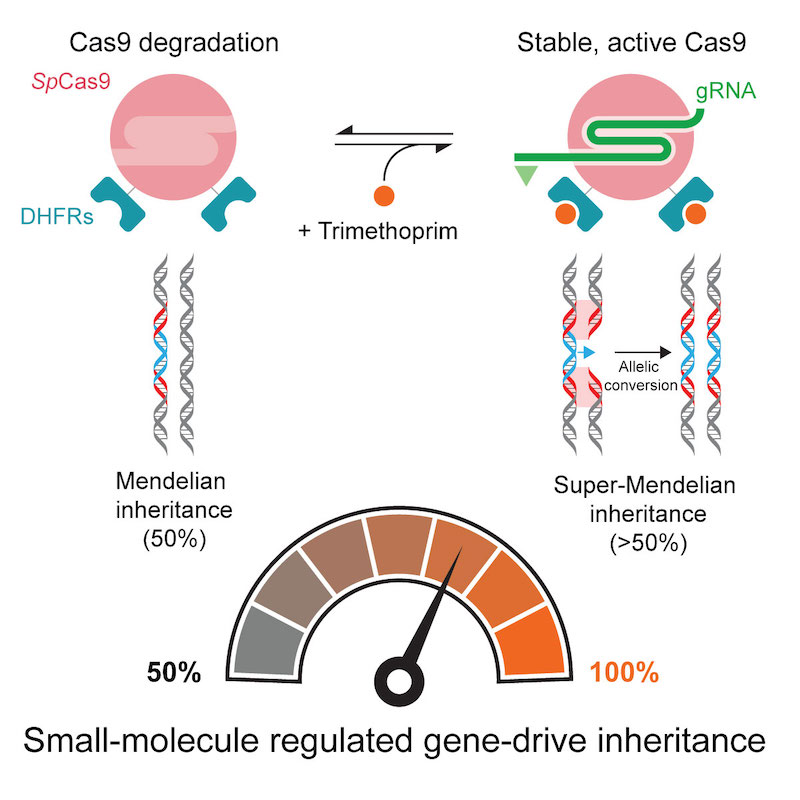
(Nat Chem Biol 2017). We fused Cas9 to destabilized domains that are largely unfolded and targeted, along with Cas9, for rapid proteasomal degradation. Addition of small molecule stabilizes these domains and rescues the fusion protein from degradation. By using this engineered Cas9 system, we were able to rapidly, reversibly, and dose-dependently control both genome editing and transcription technologies of Cas9.
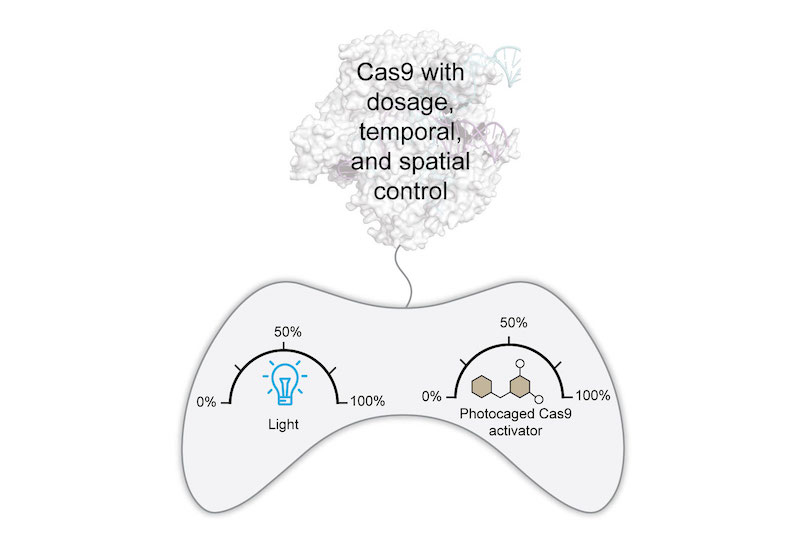
(Angew Chem Int Ed 2019). High-resolution, non-invasive, spatiotemporal control of Cas9 activity can be achieved using light. We photocaged our small-molecule activator to afford a singular system with precise dose and spatiotemporal control of Cas9 activity. This system is rapidly responsive, requires low light intensity and exposure times, and provides reversible and multi-wavelength control of Cas9.
(Cell Reports 2020). CRISPR-based gene drives have the potential to fight vector-borne diseases or suppress crop pests because they can rapidly transmit important genes through a population. They do this by surpassing the 50% inheritance limit of Mendel’s law of independent assortment through their gene-editing technology that converts a wildtype allele to a gene-drive one. However, contemporary gene drives could spread through populations to permanently modify the genome of organisms, posing safety concerns that limit their use in both laboratory and field settings if left unchecked. We describe in Drosophila the first gene-drive system controlled by an engineered Cas9 and a synthetic, orally-available small molecule, which can be applied to field applications.
Inhibitors
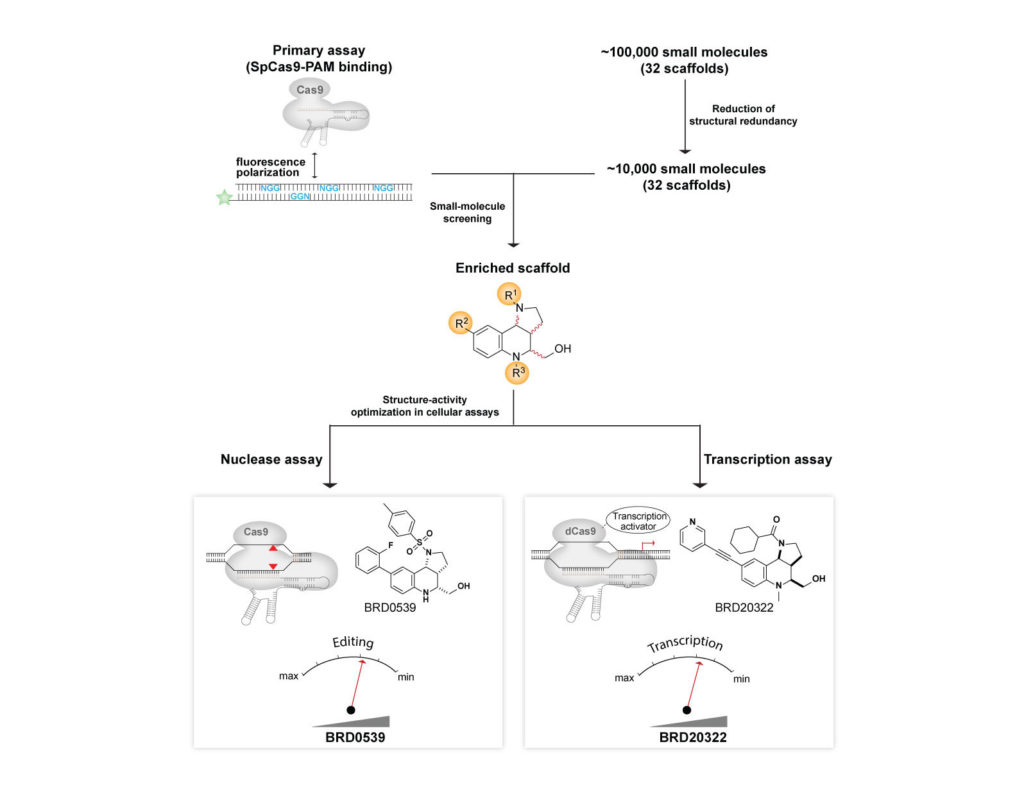
The identification of selective inhibitors of the different Cas nucleases could help improve the efficacy of gene therapies and reduce the risks associated with the off-target interactions. The identification of small-molecule inhibitors of Cas nucleases is challenging for multiple reasons. First, inhibitor identification requires robust, orthogonal, sensitive, high-throughput, miniature, and inexpensive Cas9 assays, which were unavailable. Second, Cas9 is a single turnover enzyme that holds on to its substrate with picomolar affinity throughout the biochemical reaction, adding to the challenge of developing such high-throughput assays. Third, the inhibition of Cas9 activity requires the inactivation of two nuclease domains (HNH and RuvC). Fourth, Cas9 possesses novel protein folds, limiting the ability to leverage existing rational design approaches. Fifth, Cas9 is a DNA-binding protein, a class of targets that are often deemed chemically intractable. Finally, different classes of Cas nucleases also exhibit wildly different operating mechanisms, which could make it challenging to translate assays between systems. We have resolved these issues to report first examples of cell-permeable, synthetic anti-CRISPRs.
(Chemical Sciences 2019) We have developed a sensitive, high-throughput, cell-free method wherein Cas nuclease activity is amplified via an in vitro transcription reaction that produces a fluorescent RNA:small-molecule adduct.

(Cell 2019).
We created a generalizable platform that provided the first synthetic inhibitors of Cas9 that weigh <500 Da and are cell-permeable, reversible, and stable under physiological conditions. We developed a suite of high-throughput assays for Cas9 functions, including a primary screening assay for Cas9: DNA interaction, and used these assays to screen a structurally diverse collection of natural-product-like small molecules to ultimately identify inhibitors. Using these synthetic anti-CRISPRs small molecules, we demonstrated dose and temporal control of Cas9 and catalytically impaired Cas9-based technologies, including transcription activation.
Several next-generation CRISPR-associated nucleases have emerged (e.g., SaCas9, Cas12a) with superior attributes. For example, SaCas9 is much smaller than SpCas9.19 While SpCas9 catalyzes blunt cuts, Cas12a leaves sticky ends. Cas12a employs smaller guide RNA and also can self-process an RNA sequence containing multiple gRNA target sequences into multiple gRNAs, thereby allowing easy multiplex genome editing. We have developed a suite of high-throughput assays for SpCas9, SaCas9, and Cas12a and performed small-molecule screening to identify inhibitors of these enzymes, including those that inhibit all the three enzymes (manuscripts in preparation)
Degraders
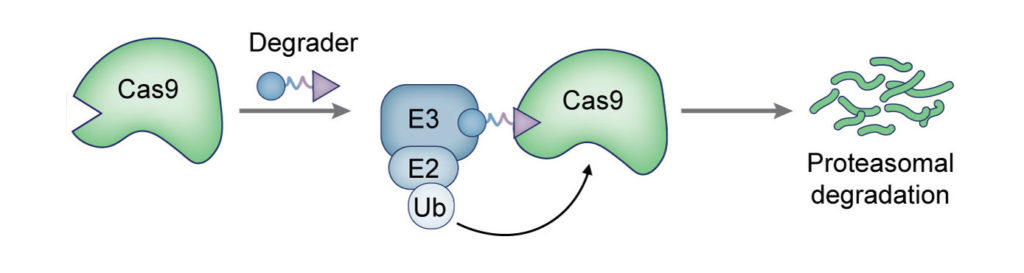
The ideal system to control half-life of CRISPR-associated nucleases and their technologies would have several characteristics. First, the system should be capable of degrading large protein:RNA complex (> 200 kDa); while SpCas9 has 1368 aa, adenine and cytosine base editors are made up of 1566 aa and 1835 aa, respectively. Base editors that can perform a simultaneous transformation of A→G and C→T are even larger, consisting of 2033 aa. Second, this system should be capable of effectively (~100 %) and rapidly (< 30 min) degrading these nucleases and their technologies to afford precision dose and temporal control as substantial editing is observed in as little as 30 min in cells. Here, a small-molecule control is preferable to other genetic methods-reduction techniques such as knockdown, which are slow to take effect. Third, the system should be minimalistic, consisting of just the controller (e.g., small molecule) and the engineered nuclease Cas9. Fourth, since CRISPR-based technologies (e.g., base editors) already bear N- and C-term fusions, genetic tags should be functional at an internal site (e.g., loops). We have developed chimeric small-molecules that can bring Cas9 in proximity to a ubiquitin ligase, enabling rapid ubiquitination and degradation of Cas9 by the proteasome. This system was able to control the Cas9 activity in a specific and dose-dependent manner, and its use ameliorated the off-target effects of the enzyme and biased the DNA repair pathways.
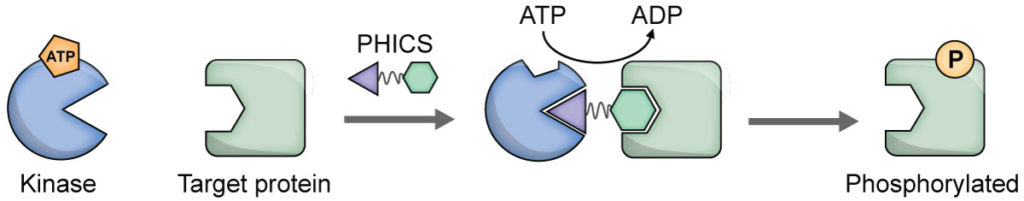
The flow of information in biological systems involves protein editing, including the addition or removal of post-translational modifications (PTMs) by “writer” or “eraser” enzymes. The disruption of homeostasis of writer/eraser enzymes underlies the pathophysiology of nearly all disorders. While CRISPR-based technologies have afforded scalable platforms for editing DNA or RNA, no such platforms exist for editing proteins. We are developing a suite of CRISPR-equivalent protein editors to add or remove PTMs by leveraging hosts’s writers/erasers. Here, we have developed a new class of editors termed Phosphorylation-Inducing Chimeric Small molecules (PHICS) that selectively induce phosphorylation of any protein-of-interest. PHICS is formed by joining a kinase binder with a binder of the target protein-of-interest so that the kinase is brought into proximity to the target protein. The resulting increase in the effective concentration of the target protein around the kinase will result in target protein phosphorylation.
The death and/or dysfunction of insulin-producing beta cells is the central pathological event in type 1 and 2 diabetes. We develop methods and design molecules for beta-cell proliferation, protection, imaging, and editing.
(J. Am. Chem. Soc. 2020). Several molecules have been identified for beta-cell proliferation, protection, and imaging. However, the lack of beta-cell specificity of these molecules jeopardizes their therapeutic potential. A general platform for selective release of small-molecule cargoes in beta cells over other islet cells ex vivo or other cell-types in an organismal context will be immensely valuable in advancing diabetes research and therapeutic development. By selectively expanding beta cells using zinc-based probes we can increase the therapeutic index of small molecules.
We leveraged the unusually high Zn(II) concentration in beta cells to develop a Zn(II)-based prodrug system to selectively and tracelessly deliver bioactive small molecules and fluorophores to beta cells. The Zn(II)-targeting mechanism enriches the inactive cargo in beta cells as compared to other pancreatic cells; importantly, Zn(II)-mediated hydrolysis triggers cargo activation. With the use of this system, we have demonstrated the selective release of multiple fluorophores in human beta cells across several cell types1, including those in pancreatic islets, and we observe selective delivery of fluorophores in vivo compared to adipose, kidney, liver, and skeletal muscle tissues. Furthermore, we have selectively induced proliferation of stem-cell-derived beta cells to allow their expansion and sorted such beta cells using this prodrug system (manuscripts under review).

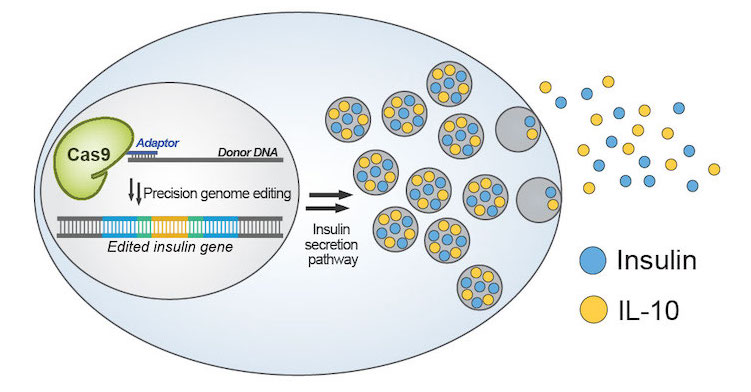
(Nature Communications) There is a diverse array of molecules that can improve the efficiency and outcome of Cas9-based editing. We have developed a platform for conjugating these molecules to Cas9 to increase their local concentration around the editing site. This platform allows for the site-specific and multiple-site conjugation of a wide assortment of molecules on both the termini and internal sites of Cas9, making it applicable for countless gene-editing applications. Using this platform, we modified a Cas9 with multiple single-stranded oligonucleotide donor (ssODN) to more precisely incorporate it at the DNA break site—significantly increasing the efficiency of precision genome editing.
Current β-cell transplantation therapies for type 1 diabetes suffer from immune rejection, resulting in acute cell loss and only short-term therapeutic effects. The macroencapsulation of β cells with a semipermeable membrane can protect them from the host’s immune system, though foreign body reaction-induced fibrosis can impair the mass transfer and viability of encapsulated cells. Anti-inflammatory cytokines, such as interleukin 10 (IL-10), can reduce fibrosis and promote long-term β-cell survival and superior islet function. Therefore, engineered β-cells that secrete anti-inflammatory cytokines and anti-fibrotic factors could propel the development of cell-based therapeutics for diabetes. By leveraging the ssODN conjugation platform, we successfully engineered a β-cell line to repurpose the insulin secretion machinery. This enabled the glucose-dependent secretion of protective immunomodulatory factor interleukin-10 to protect the cells from damage.
Nature has evolved organisms that feed frequently (e.g., humans) and those that feed infrequently (e.g., lions). While frequent feeding organisms are often studied (e.g., mice), it is the infrequent feeding organisms that survive conditions considered pathological to humans. We are unraveling the molecular mechanisms by which infrequent feeding reptiles avert metabolic disorders despite lifestyles that would kill humans. We draw inspiration from the studies on the infrequent-feeding Gila monster that led to the diabetes drug exenatide. See the New York Times article here for more details.